The Effect of Metal Ions of Varying Sizes and Concentrations on DNA Melting Temperature
George Schank
Abstract
As DNA is heated in solution, it begins to denature. The native double helix form of DNA dissociates into single strands (melting). Many factors affect the temperature at which DNA transforms from a double stranded form to a single stranded form. The presence of cations is one such factor that can have a stabilizing affect on the double helix form of DNA.
The size, charge, and concentration of the cation in solution are key factors that effect the stabilization of DNA. This experiment tested each of these variables with respect to ions having different size and charge (K+, Mg2+, Ca2+, Ba2+, and Zn2+) in order to determine the ion with the most effective combination of charge and size that increases the melting temperature. This was accomplished by altering one variable while the other remained constant.
It was determined that potassium and barium had no effect on the melting temperature, while magnesium and calcium increased the melting temperature. Furthermore, magnesium outperformed calcium and was found to be the most effective ion at higher concentrations for stabilizing DNA.
Introduction
DNA (deoxyribonucleic acid) is a nucleic acid that carries genetic information and has a double helix structure. It is made up of a chain of nucleotides which contains a sugar, a phosphate, and a base. The four variations of nucleotides are: adenine, thymine, guanine, and cytosine. Adenosine and thymine bind together (with two hydrogen bonds) and guanine and cytosine bind together (with three hydrogen bonds). This is significant when the stability of the double-helix is examined. The H-bonding between the bases must be overcome if the DNA is to �melt� from its double-helix form into a single strand. There are many factors which can influence the overall stability of DNA, such as G-C content, chain length, and environment (temperature, pH, presence of ions, etc.).
It is common knowledge that the higher the content of guanine and cytosine bonds, the more stable the DNA. This is due to G-C bonds having three hydrogen bonds instead of only two, like A-T bonds. It was also suggested that higher cation concentrations will behave in the same manner and stabilize the DNA by forming a shell around the negatively charged phosphates of DNA. This would potentially result in proportionally higher melting temperatures, depending on the concentration (Resendis-Antonio, 2003). However, the effects of cations of differing sizes and concentrations have not been extensively compared. It is reasonable to expect that the larger the cation, the less extensive the effect it will have on the DNA since larger ions are not expected to cluster around the backbone as extensively as the smaller cations.
Denaturation is a process in which a molecule undergoes a transformation and is no longer in its native state. This can be caused by heat, acids, bases, detergents, or other specific chemicals. When heat is the source of denaturation, it is, in the case of DNA, associated with the melting temperature. DNA�s melting temperature (Tm) can be defined as the temperature at which half of the DNA molecules in a solution have dissociated into single strand molecules.
There have been a number of studies conducted in the past to determine the melting point of DNA. For instance, it had been previously established by an earlier study by Owczarzy, et al, that the sodium ion stabilizes the backbone structure of DNA (Owczarzy, et al, 2004). It stabilizes the backbone because the phosphate groups are moderately strong acids, and thus they are almost completely ionized at physiological pH, which allows interactions between the backbone and the cations. Stabilization of the backbone affects the temperature at which the DNA denatures (i.e. the more the ionic interaction, the more stable the DNA and the higher the melting temperature). Many experiments have been performed involving DNA, its melting temperature, and the factors that affect the Tm. However, availability of different approaches indicates that there is still much to explore concerning this type of experiment.
This experiment investigates the effects of potassium, magnesium, calcium, barium, and zinc ions on the stability of the double-helix. The results of this study potentially may lead to more types of applications in clinical, medicinal, pharmaceutical, and forensic research, as well as in genetic engineering.
DNA has a maximum absorbance at a wavelength of 260 nm. Single stranded form has a higher molar absorptivity than the double stranded form at the same wavelength. In this study, this difference in absorptivities is used to measure the melting temperature by graphing the change in absorbance as the double stranded DNA denatures to single stranded DNA with increasing temperature.
Materials and Methods
Materials:
-Herring sperm DNA from Promega
-size range: 100-3000bp (for at least 80% of the DNA fragments)
-concentration: 10 mg/mL (original concentration)
-Calcium chloride, potassium chloride, zinc chloride, barium chloride, and magnesium chloride (all reagent grade)
-Equipment: SP 2000 UV-Vis, Thermo-Spectronic heating sleeve, Thelco water bath (model 85), pump
Procedure:
The absorbance of the standard solution of DNA (0.008 mg/�L) at 260 nm was measured using UV-Vis spectrophotometer at different temperatures ranging from 25o to 100o C.
The DNA-metal ion solutions tested contained 0.02 mg of DNA and different concentrations (10-4, 10-3, or 10-2 M) of metal ions. Absorbance measurements were done using a blank solution of the respective metal ion concentration at 260 nm, at the temperature range of 25o to 100o C.
Experimental Data and Results
The initial concentration of the metal ion ranged from 10-4 M to 10 -2 M. The concentration of DNA used for this comparison was 0.008 mg/�L due to the known linearity of the standard curve at low concentrations.
Table 1: Absorbances of DNA solutions without metal ion at 260 nm
|
Temp., C |
Abs, 260 nm |
|
25 |
0.107 |
|
30 |
0.108 |
|
35 |
0.109 |
|
40 |
0.109 |
|
45 |
0.112 |
|
50 |
0.115 |
|
55 |
0.117 |
|
60 |
0.121 |
|
65 |
0.130 |
|
70 |
0.138 |
|
75 |
0.138 |
|
80 |
0.136 |
|
85 |
0.138 |
|
90 |
0.138 |
|
95 |
0.137 |
|
100 |
0.137 |
Graph 1: Absorbances of DNA solutions without metal ion at 260 nm

Table 2: Absorbances of DNA solution containing K+ ions at 260 nm
|
Temp., C |
10 -2 M |
10-3 M |
10-4 M |
|
25 |
0.109 |
0.095 |
0.114 |
|
30 |
0.112 |
0.097 |
0.118 |
|
35 |
0.115 |
0.100 |
0.122 |
|
40 |
0.119 |
0.105 |
0.125 |
|
45 |
0.125 |
0.107 |
0.127 |
|
50 |
0.128 |
0.110 |
0.128 |
|
55 |
0.131 |
0.112 |
0.128 |
|
60 |
0.135 |
0.115 |
0.130 |
|
65 |
0.138 |
0.116 |
0.131 |
|
70 |
0.137 |
0.118 |
0.135 |
|
75 |
0.140 |
0.120 |
0.139 |
|
80 |
0.145 |
0.125 |
0.139 |
|
85 |
0.145 |
0.125 |
0.140 |
|
90 |
0.146 |
0.128 |
0.141 |
|
95 |
0.147 |
0.129 |
0.142 |
|
100 |
0.148 |
0.129 |
0.142 |
Graph 2: Absorbances of DNA solution containing K+ ions at 260 nm
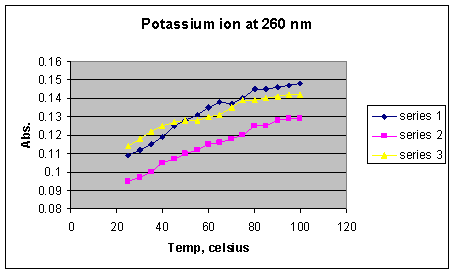
*Series 1 = 1 x 10-2 M K+; Series 2 = 1 x 10-3 M K+; Series 3 = 1 x 10-4 M
Table 3: Absorbances of DNA solution containing Mg2+ ions at 260 nm
|
Temp., C |
10 -2 M |
10-3 M |
10-4 M |
|
25 |
0.128 |
0.119 |
0.110 |
|
30 |
0.130 |
0.120 |
0.111 |
|
35 |
0.133 |
0.122 |
0.112 |
|
40 |
0.135 |
0.124 |
0.114 |
|
45 |
0.136 |
0.125 |
0.115 |
|
50 |
0.141 |
0.130 |
0.118 |
|
55 |
0.142 |
0.134 |
0.122 |
|
60 |
0.145 |
0.138 |
0.125 |
|
65 |
0.147 |
0.141 |
0.128 |
|
70 |
0.151 |
0.145 |
0.134 |
|
75 |
0.158 |
0.149 |
0.139 |
|
80 |
0.162 |
0.152 |
0.142 |
|
85 |
0.166 |
0.157 |
0.144 |
|
90 |
0.167 |
0.158 |
0.146 |
|
95 |
0.168 |
0.159 |
0.146 |
|
100 |
0.167 |
0.157 |
0.146 |
Graph 3: Absorbances of DNA solution containing Mg2+ ions at 260 nm
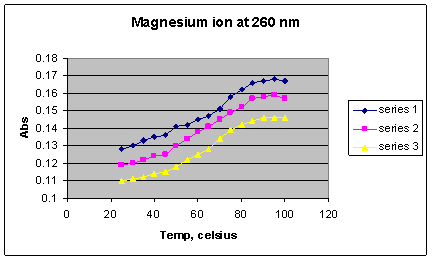
*Series 1 = 1 x 10-2 M Mg2+; Series 2 = 1 x 10-3 M Mg 2+; Series 3 = 1 x 10-4
Table 4: Absorbances of DNA solution containing Ca2+ ions at 260 nm
|
Temp., C |
10 -2 M |
10-3 M |
10-4 M |
|
25 |
0.089 |
0.098 |
0.099 |
|
30 |
0.090 |
0.100 |
0.101 |
|
35 |
0.091 |
0.100 |
0.103 |
|
40 |
0.092 |
0.103 |
0.105 |
|
45 |
0.094 |
0.104 |
0.105 |
|
50 |
0.096 |
0.106 |
0.107 |
|
55 |
0.101 |
0.115 |
0.115 |
|
60 |
0.105 |
0.120 |
0.122 |
|
65 |
0.108 |
0.126 |
0.125 |
|
70 |
0.110 |
0.130 |
0.132 |
|
75 |
0.113 |
0.135 |
0.134 |
|
80 |
0.114 |
0.137 |
0.138 |
|
85 |
0.113 |
0.140 |
0.137 |
|
90 |
0.112 |
0.141 |
0.139 |
|
95 |
0.113 |
0.140 |
0.138 |
|
100 |
0.113 |
0.141 |
0.138 |
Graph 4: Absorbances of DNA solution containing Ca2+ ions at 260 nm
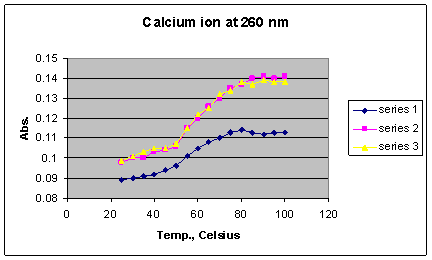
*Series 1 = 1 x 10-2 M Ca2+; Series 2 = 1 x 10-3 M Ca2+; Series 3 = 1 x 10-4 M Ca2+
Table 5: Absorbances of DNA solution containing Ba2+ ions at 260 nm
|
Temp., C |
10 -2 M |
10-3 M |
10-4 M |
|
25 |
0.115 |
0.133 |
0.120 |
|
30 |
0.117 |
0.133 |
0.123 |
|
35 |
0.120 |
0.133 |
0.125 |
|
40 |
0.125 |
0.140 |
0.128 |
|
45 |
0.133 |
0.144 |
0.132 |
|
50 |
0.140 |
0.149 |
0.135 |
|
55 |
0.145 |
0.155 |
0.138 |
|
60 |
0.150 |
0.160 |
0.142 |
|
65 |
0.154 |
0.163 |
0.147 |
|
70 |
0.157 |
0.168 |
0.151 |
|
75 |
0.159 |
0.173 |
0.153 |
|
80 |
0.161 |
0.174 |
0.155 |
|
85 |
0.161 |
0.175 |
0.158 |
|
90 |
0.161 |
0.176 |
0.162 |
|
95 |
0.159 |
0.178 |
0.161 |
|
100 |
0.159 |
0.178 |
0.161 |
Graph 5: Absorbances of DNA solution containing Ba2+ ions at 260 nm
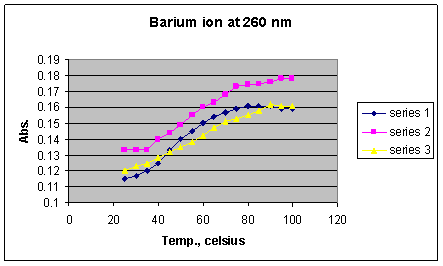
*Series 1 = 1 x 10-2 M Ba2+; Series 2 = 1 x 10-3 M Ba2+; Series 3 = 1 x 10-4 M Ba2+
Table 6: Absorbances of DNA solution containing Zn2+ ions at 260 nm
|
Temp., C |
10 -2 M |
10-3 M |
10-4 M |
|
25 |
0.060 |
0.089 |
0.088 |
|
30 |
0.059 |
0.095 |
0.091 |
|
35 |
0.060 |
0.100 |
0.094 |
|
40 |
0.059 |
0.103 |
0.097 |
|
45 |
0.061 |
0.105 |
0.099 |
|
50 |
0.055 |
0.108 |
0.099 |
|
55 |
0.050 |
0.108 |
0.103 |
|
60 |
0.048 |
0.108 |
0.105 |
|
65 |
0.040 |
0.107 |
0.105 |
|
70 |
0.033 |
0.103 |
0.102 |
|
75 |
0.027 |
0.102 |
0.100 |
|
80 |
0.026 |
0.103 |
0.099 |
|
85 |
0.02 |
0.102 |
0.098 |
|
90 |
0.021 |
0.102 |
0.099 |
|
95 |
0.020 |
0.103 |
0.099 |
|
100 |
0.019 |
0.102 |
0.100 |
Graph 6: Absorbances of DNA solution containing Zn2+
ions at 260 nm 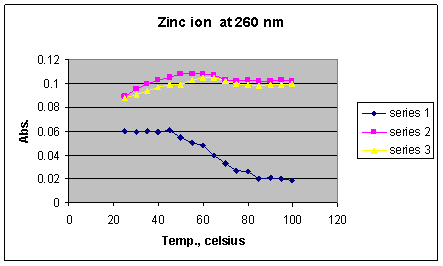
*Series 1 = 1 x 10-2 M Zn2+; Series 2 = 1 x 10-3 M Zn 2+; Series 3 = 1 x 10-4 M Zn 2+
Conclusion
The phosphodiester backbone of DNA contains, as the name suggests, phosphate groups. These phosphate groups alternate with sugar rings, forming the backbone of the DNA molecule. The phosphates are located on the outside of the DNA, as shown in the picture below:
*In the picture, the phosphate groups are seen as yellow, while the oxygen atoms (four per phosphate) are seen as red.
The structure of DNA is stabilized by internal hydrogen bonds between purines and pyrimidines. Externally, electronegative oxygen atoms have the potential to form hydrogen bonds with surrounding molecules. The positioning of the phosphate groups prevents them from having a significant effect on each other; however, they do still have some slight repulsion in-between. Their positioning on the exterior of the DNA molecule allows them to interact with cations, which are able to lessen the repulsion between the phosphate groups. The result is a more stable DNA molecule with a higher overall melting temperature when it is in a solution containing cations. In other words, the two strands of DNA are both negatively charged, and, hence, are repelled by one another due to the large charge density. The addition of positively charged ions can reduce the charge density by surrounding and interacting with the negative charges, thereby stabilizing the molecule.
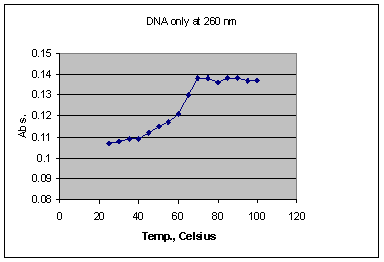
Based on the results of this experiment, it appears that a solution of DNA only (Graph 1) denatures over a temperature range of 40 to 70o C, with a melting temperature of 65o C, and a total change in absorbance of 0.025. The large range of denaturation temperature is attributed to the fact that the DNA used was not comprised of all same-length strands. Hence, the shorter chains denatured at lower temperatures due to a lower overall bond strength caused by fewer hydrogen bonds, assuming the effect of hydrogen bonding on stability is cumulative.
Effect of K+ ion:
In the case of the potassium ion (Graph 2), it appears that the bulk of the DNA denatures at approximately the same temperature (30 to 80 o C) for all concentrations of ion, with an average Tm of 65o C (from the midpoint of the steepest portion of the melting curves). Due to the larger range of denaturation temperatures than that of DNA alone, as well as no net difference of effect between the different concentrations, it appears that potassium has no effect on DNA stabilization in solution. The results seem to indicate that the other doubly charged ions may be able to stabilize more than one phosphate group, whereas potassium can not.
Effect of Mg2+ ion:
Graph 7: Melting temperatures at three different concentrations of magnesium ion
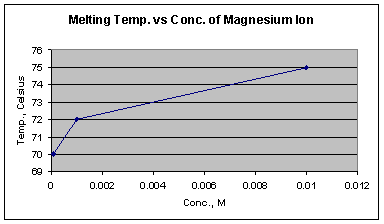
Graph 7 shows that as the concentration of the magnesium ion increases, the Tm increases, indicating that higher concentrations are better able to stabilize the double stranded DNA. The melting temperatures of magnesium samples (Graph 3) at all concentrations had increased by approximately 8o C, with an average Tm of 72.67o C (from the midpoint of the steepest portion of the melting curves), and the highest Tm being 76 o C at the concentration of 10-2 M. This shift in melting temperature is the largest effect caused by any of the ions.
Effect of Ca2+ ion:
Graph 8: Melting temperatures at three different concentrations of calcium ion
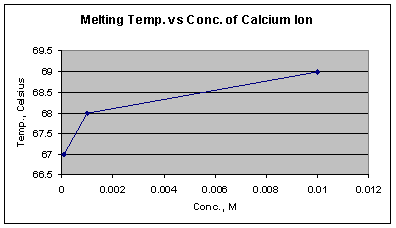
Graph 8 shows that as the concentration of the calcium ion increases, the Tm increases, indicating that higher concentrations are better able to stabilize double stranded DNA. The change in melting temperature is greater between 10-4 M and 10-3 M than between 10-3 M and 10-2 M. This trend which also is observed with Mg2+ ion implies that after a certain metal ion concentration is reached, a kind of saturation effect results relative to the maximum possible number of ions spatially surrounding the phosphate groups. The calcium ion seemed to have the second largest effect on the DNA�s stability (Graph 4) with an average Tm of 68o C and the highest Tm being 69 o C at the concentration of 10-2 M.
Effect of Ba2+ ion:
The barium ion (Graph 5) displayed a denaturation temperature range of 40 to 80o C, with an average Tm of 65o C (from the midpoint of the steepest portion of the melting curves), and an ending absorbance of 0.114, both of which are similar to the DNA sample containing no ions. Therefore it can be concluded that this ion has no real effect on stability. The lack of effect is suspected to be due to the barium ion being too large to allow for multiple ions to form an effective density of positive ions around the negatively charged backbone.
Effect of Zn2+ ion:
Results of the zinc samples (Graph 6) indicate that as DNA begins to denature, the absorbance drops dramatically. This observation suggests the possibility of a complex formation between the zinc ion and single-stranded DNA, resulting in the depletion of single-stranded DNA which absorbs at 260 nm. Since the sample with the highest concentration of Zn2+ had the largest drop in absorbance, the complex was most stable at this concentration, indicating a higher amount of complex formation than the other two concentrations. Furthermore, complex formation appears to be occurring in the temperature range of 45 to 80o C, the same range in which the DNA denatures, indicating that the complex formation increases with temperature.
One possible explanation is that zinc is present in proteins which are involved in binding DNA. This explanation is supported by the existence of zinc fingers containing Zn2+ ions in the protein domain (Junker 2006), which promote protein-DNA interactions. Despite these interesting findings, the stabilizing effect of zinc was not observed.
Table 7: Comparison of effect of ions (at their most effective concentration) on melting temperature of DNA
|
Ion |
Conc., M |
Tm, o C |
|
K+ |
10-3 |
66 |
|
Mg2+ |
10-2 |
76 |
|
Ca2+ |
10-2 |
69 |
|
Ba2+ |
10-4 |
65 |
|
Zn2+ |
10-4 |
N/A |
*Tm of DNA solution without a metal ion: 65o C
*For Zn2+, measurement of Tm could not be done due to loss of absorbance of single stranded DNA via coordination with Zn2+ ions
The results of this experiment clearly indicate that magnesium and calcium had the most stabilizing effect on double stranded DNA. Magnesium had the largest effect on melting temperature, with a Tm of 76oC at a concentration of 10-2 M, compared to 65 o C for DNA alone. DNA solution containing Ca2+ at a concentration of 10-2 M had a Tm of 69o C, which was higher than that of DNA alone. Magnesium and Calcium were both the ideal size and charge to provide the maximum amount of stabilization, which resulted in a higher temperature to convert 50% of double-stranded DNA to single stranded DNA. However, since magnesium is smaller than calcium, the Mg2+ ions were better able to cluster around the backbone, providing more stabilization, resulting in a higher Tm. Hence, magnesium, more so than all other ions tested, has the largest potential for impact on DNA research by providing a more stable molecule.
References
Bayer, J.; Radler, J. O.; Blossey, R.. "Chains, Dimers, and Sandwiches: Melting Behavior of DNA Nanoassemblies". Nano Lett.; (Communication);2005; 5(3); 497 � 501.
Becaud, J.; Pompizi, I.; Leumann, C. J.. �Propagation of Melting Cooperativity along the Phosphodiester Backbone of DNA� J. Am. Chem. Soc; 2003. 125(50); 15338-15342.
Coene, Marc; Cocito, Carlo. �A microanalytical procedure for determination of the base composition of DNA.� European Journal of Biochemistry. 1985; 150(3); 475.
Dorte Renneberg and Christian J. Leumann. �Watson-Crick Base-Pairing Properties of Tricyclo-DNA." J. Am. Chem. Soc.; (Article); 2002; 124(21); 5993-6002.
Junker, Matthew. Matthew Junker, Assistant Professor. University of Virginia. 12 Apr. 2006 <http://nsm1.utdallas.edu/bio/Junker/junker.htm>.
Lixiang Sun and Yuxiang Bu. �Marked Variations of Dissociation Energy and H-Bond Character of the Guanine-Cytosine Base Pair Induced by One-Electron Oxidation and Li+ Cation Coupling" J. Phys. Chem. B. ; (Article); 2005; 109(1); 593-600.
Owczarzy, Yong You, Bernardo G. Moreira, Jeffrey A. Manthey, Lingyan Huang, Mark A. Behlke, and Joseph A. Walder."Effects of Sodium Ions on DNA Duplex Oligomers: Improved Predictions of Melting Temperatures." Biochemistry ; (Article); 2004; 43(12); 3537-3554.
Resendis-Antonio, O.; Garcia-Colin, L.S.; Larralde, H.. Physica A.. �A statistical model of DNA denaturation�. Physica A; 2003; 318(3);435.
Sato, Yu-ichi; Kobayashi, Yuki; Kamiya, Takayuki; Watanabe, Hiromitsu; Akaike, Toshihiro; Yoshikawa, Kenichi; Maruyama, Atsushi. Biomaterials. �The effect of backbone structure on polycation comb-type copolymer/DNA interactions and the molecular assembly of DNA.� 2005; 26(7); 703.
Sau Lawrence Lee; Pablo G. Debenedetti; Jeffrey R. Errington; Brian A. Pethica; and David J. Moore. "A Calorimetric and Spectroscopic Study of DNA at Low Hydration� J. Phys. Chem. B.; (Article); 2004; 108(9); 3098-3106.
*Special thanks to Dr. Ozturk, Mr. Stratmann, and Dr. Henshaw for their assistance throughout the experiment.
©